Life sentence: teaching large language models to read the book of life

Now you’re talking…
If you’ve used one of the recent high-profile AI chatbots, then you’ve no doubt been wowed by their capability. If you haven’t used one, then you’re in an increasingly small population, as these large language models have attracted enormous publicity.
A lot of the publicity has focussed on how good the models are at crafting text. There have been endless articles, reports and segments on news programs where the ‘big reveal’ at the end is that the whole article or script has been written by an AI chatbot[1]. Similarly, we’ve seen a few examples of the machines being ‘caught out’ producing factually incorrect content which have then gone viral, often with some ‘here’s why the machines will not be taking your job’ type framing.
These systems (and you can insert the names of any of your favourite high-profile large language models here) all use a similar construction, based around the tokenisation of inputs and a ‘transformer’ architecture. Their core function is to predict the next word given a particular text prompt – one word at a time. Longer passages of text are generated simply by feeding previous output back into the system as part of the input. In many of these AI chatbots, you can see this happening in real time as each word is printed out as if typed by an intelligent stranger. All fairly straightforward, but strange things start to happen as these models become larger and more training data (and more compute resources) are put into training the system. The systems start to exhibit additional abilities that we might not expect to be present in a system that, at its heart, is a statistical engine which is surfacing the most probable next word.
Emergent properties
As the systems grow larger in terms of the number of trainable parameters, and as more training data is fed in, and consequently more computing resources are required to train these larger models on larger datasets, the systems start to exhibit new abilities. Even with relatively smaller systems, systems start to exhibit question answering ability. For the largest models, the systems exhibit qualities such as logic and reasoning, mathematical abilities, theory of mind, spatial reasoning[2]. These ‘emergent properties’ mean that large models produce very different and qualitatively far better output for a given prompt than smaller models.
Why might these emergent properties exist though? Those at the frontiers of AI research do not have a single clear answer for why we see these emergent properties as AIs ‘size’[3] increases. One theory that makes instinctive sense to the authors has been advanced by Ilya Sutskever, Chief Scientist at (and co-founder of) OpenAI. Sutskever has argued that emergent properties are a necessary result of the task of predicting the next word, as in order to perform that prediction task more perfectly, a machine must derive an understanding of the reality described in the various materials used in training.[4]

From syntax to semantics
This is genuinely remarkable, and perhaps even more so when we consider how such systems are trained. In crude terms, training of large language models is done by showing them short snippets of text with a missing word, asking the system to guess the word, and then tweaking the parameters of the model depending on how far from correct the system’s guess is.[5]
As a result, the process of building and training a large language model is one that allows our machines to derive real semantic meaning, and encode an understanding of the world in their weights and biases, having only been exposed to snippets of ‘fill in the blank’ exercises. We feed it hundreds of examples of form, and the machine gradually starts to understand function.
The only choices that model designers make when building an LLM that would in any way code for semantic context is which collections of letters are identified as ‘words’ (technically identified as ‘tokens’, which might be part words, common suffixes etc.) in the ingestion or word encoding layer. The machine is otherwise only exposed to text with no other assistance in understanding, and must derive that understanding for itself.
Given the emergent properties that are then observed once this process is repeated a few trillions of times, this system has proved itself capable of taking letters, words and sentences and developing a real understanding of our human-scale world.
The (amino) acid test
Are human languages the only data structures that could be used with LLMs? Certainly not, and we have already seen transformer models with tokenised inputs used for everything from playing video games to controlling robots. Most of these are explorations of human created content (games, music, art etc.) or explorations of the human-scale world but from a different perspective to written text (robot control, self-driving vehicles, etc.).
What if we took something at a different scale altogether? We often hear aspects of biology being referred to as the ‘book of life’. Can we take that metaphor more literally?
In a traditional LLM, we see a structure of:
- letters (plus numbers, punctuation, etc.); forming
- words (or more accurately tokens, since suffixes, punctuation marks, etc. are often tokens in their own right); forming
- sentences and paragraphs.
In our LLM repurposed for biology, we might start with amino acids as the smallest unit, taking the role of letters, with various ions acting as ‘punctuation’. We then use our amino acid ‘letters’ to form protein ‘words’. Finally, these protein ‘words’ form biological pathways made up of many protein interactions, and are the ‘sentences’ or longer prose of our ‘Large Bio-Chemistry Model’ or LBCM.
We can then work out the relative information content of each domain for our LLM and LBCM, and therefore extrapolate how big an LBCM might need to be for us to expect to see emergent properties.
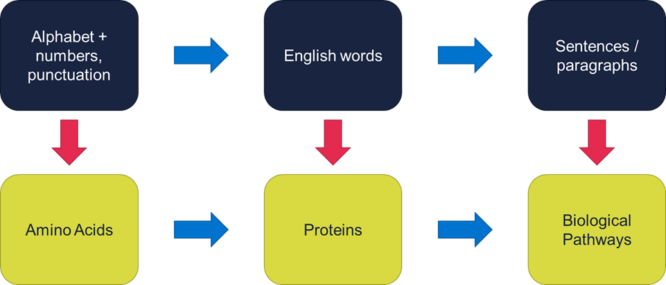
Figure 2 – Mapping language features to biochemistry features.
Comparing sizes
A fair warning for those who have read this far: the article is about to turn from metaphor to more specific technical explorations of our biochemical LLM. If you want to skip past the numbers, pick up the article further down at the section headed “Structural and data challenges”.
In an LLM targeting the English language, there are 26 letters of the alphabet. However, we need to double this to allow for capitals, and then add a few hundred other characters, including numbers, punctuation marks and letters with diacritical marks. The average word length in English is between 5 and 6 letters, with the longest words topping out at a few tens of letters for most purposes (antidisestablishmentarianism, anyone?), although some rarely-encountered scientific terms contain hundreds or even thousands of letters[6]. Someone who is fluent in English might have a vocabulary of around 8,000 to 10,000 words, which will include a variety of proper nouns from popular culture. Working out average sentence length is more difficult, and greatly depends upon the context and subject matter of the text, and the writing style of the author. In this paper the average sentence length is just over 18 words, so we’ll round up to 20 words for this calculation.
All well and good, but while the largest LLMs might target the English language, in many cases they have been exposed to tens of other languages. The latest models have been tested to be capable of sensibly responding in somewhere between 25 and 90 languages, depending on the tests[7]. Many languages have a smaller working vocabulary than English, but it would be sensible to allow for around 3,000 words per language on average. For 90 languages this would result in a total vocabulary of 270,000 words. Our character set is also proportionately larger to allow for the alphabets and pictograms used by all of these additional languages. Indeed most language models rely on the UTF-8 character encoding standard, with its just over 1 million characters.
How do these numbers compare to the parametric capacities and training data set sizes of the largest LLMs? Multiplying a 270,000 word vocabulary, by 20 words per sentence, the result is 5,400,000 combinations. Since punctuation marks, numbers and prefixes and suffixes of words might be tokenised separately, 20 words might encode as 30 ‘tokens’, so that takes us up to 8,100,000 combinations. However, most LLMs have an input limit on tokens that is much higher than a single sentence, with many medium-scale models allowing for an input of 2,000 or so tokens[8]. That takes the potential combinations of tokens to around 540,000,000, although the vast majority of those combinations would be gibberish. With the most performant LLMs currently having between 150 billion and 1 trillion trainable parameters, the ratio of parameters to character combinations is around three orders of magnitude greater, or a ratio of 1,000:1. Current state-of-the-art LLM training methods tend to want around 10 to 20 training tokens per trainable parameter, so the size of the dataset needed is around 300 billion to 2 trillion words.
Looking at the biochemistry side of things, how do things compare? Starting with our letter/character equivalent, there are 21 proteinogenic amino acids (i.e. amino acids that form proteins)[9]. In addition to these 21 amino acid ‘letters’, we would need to make some allowance for stray ions that might be present in the environment. Without numbers, punctuation marks or diacritical marks, the size of our LBCM ‘character’ set will be potentially smaller than for an LLM, and 100 or so ‘characters’ might suffice. In terms of our protein ‘word’ length, amino acid combinations of less than 50 or so amino acids tend to be classed as peptides[10], with chains of 50 or more being proteins. Studies have shown the average human protein chains to be around 375 amino acid long, with the largest topping out at over 34,000[11] amino acids. Our LBCM protein vocabulary also needs to be larger, with at least 20,000 different ‘words’ to be tokenised[12]. These proteins then interact in biological pathway ‘sentences’. While there are a large number of these pathways that have been mapped out, few are of significant length[13]. Therefore the same 2,000 token input limit would seem to be more than sufficient.
Multiplying that out, if we took a 2,000 token limit, by a 20,000 protein ‘word’ vocabulary, then our LBCM would have about 4,000,000 input combinations, which is much smaller than the numbers for the largest LLMs above. Assuming that the same ratios of trainable parameters and training tokens to input combinations are needed to start to see emergent properties[14], our LBCM should start to exhibit these properties at sizes equivalent to the larger LLMs.[15]
| Attributes | Single Language LLM | Large BioChem Model | Largest LLMs |
| ‘Letter’ layer | 128 ASCII characters | 21 amino acids (proteinogenic amino acids) | 1m+ UTF-8 characters |
| ‘Word’ layer | ~10,000 words | ~20,000 proteins | ~200,000+ Words across 90 languages |
| ‘Word’ layer length | Avg. 5 – 6 letters, longest a few tens of letters | Avg. 350 – 400 amino acids, longest 34,350 amino acids | On average 4 glyphs (many languages use single pictograms/word) |
| ‘Sentence’ length | Highly variable – LLMs typically allow input of over 1,000 words. | Biological Pathways relatively short, but many chain together. Allow a similar 1,000 to 2,000 ‘tokens’. | Larges LLMs allow input of up to 25,000 ‘tokens’, or roughly 17,000 words |
| Model sizes | 5 to 15 billion parameters | Useful model size TBC | 200 billion to 1 trillion+ parameters |
Structural and data challenges
There is certainly a larger information space to be explored for the LBCM than an LLM. If the structure were similar, with a protein encoding layer (the equivalent of a word encoding layer in an LLM) vectorising inputs before self-attention layers find further semantic connections between inputs, it is interesting to consider what properties the protein encoding layer might encode.
With a typical word encoding layer, we can directly explore some semantic relationships represented within the word vectors. It becomes possible to see how words with similar semantic context cluster together. With a protein encoding layer, we might expect that protein-specific features would be encoded within that space, with 3D structure, binding points, charge(s) at different points on the surface, etc. presumably all being represented. In effect, the protein encoding layer might need to learn as much information as earlier protein-folding specific AI models, meaning that an LBCM might need to rebalance the distribution of trainable parameters away from self-attention layers and toward the encoding and decoding layers.
Another challenge to consider is the potential difficulties with sourcing data. Where repositories of terabytes of text from various sources already exist for training LLMs, sourcing the quantity of data required to train an LBCM using a similar approach may prove more challenging. While there are open repositories of such data, testing and training an LBCM would require a far greater investment in data collection and output testing. However, given the likely investment needed in compute resources to train a hundred-billion plus parameter machine learning model, whether these additional hurdles create a significant further barrier for entry to this area is debateable.
What kind of emergent properties might we expect?
At the risk of tautology, what unexpected properties might we expect to emerge? If Sutskever’s ideas[16] about why word prediction engines start to manifest reasoning, mathematics, etc., are correct, then these emergent properties are a necessary function of the model’s need to ever more perfectly represent of the reality of the world in order to provide a better prediction. Therefore any LBCM emergent properties would stem from its steadily better representation of the reality of biochemistry. We might expect that such systems could provide hyper-personalised outcomes, or build protein bio-machines capable of extremely targeted treatments, destroying particular proteins, or repairing others. In effect, we might expect these systems to be capable of reasoning about, and generating meaningful biochemical sequencing in the same way that an LLM can reason about and generate meaningful text.
The real power of such systems might best be unlocked if combined with a traditional LLM. If LLMs can already address multiple languages, then can we add speaking ‘protein’ to that list? This pairing would then provide such a system with an interface where natural language queries regarding particular biological processes could be queried, and the resulting model might be able to provide outputs both in terms of the protein interactions and a written explanation of the reasoning underpinning the process.

Comparing the LBCM to LLM from a legal risk perspective
Any biologically-targeted large model would present a different risk profile to other LLMs. It is extremely likely that such systems would constitute software as a medical device for the purposes of relevant regulatory regimes. As such it would need to be subject to conformity assessment. In addition, an AI that itself constitutes a product subject to the CE marking regime would then be considered a ‘high risk’ AI use case under the forthcoming EU AI Act (‘AIA’).
However, since training data sets for the biological pathways could be derived from public research data, and none of it constitutes ‘personal data’ from the perspective of common privacy law definitions, training an LBCM might present a much lower legal risk surface from an intellectual property and data protection perspective than is sometimes the case in relation to LLMs. Similarly, any LBCM would be unlikely (on its own) to constitute a ‘general purpose’ AI for the purposes of the AIA, and therefore the provisions relating to general purpose AI in that Act would not be triggered. That said, an LBCM that is structured as an ‘extra language’ spoken by an LLM would still be caught by that definition, and since that form of deployment is likely to be of the greatest utility, the general purpose AI provisions would then need to be considered during training, testing and deployment.
Next steps
There are clearly enormous opportunities to use LLM structures for pharmaceutical use cases. Scanning literature and pulling out previously unnoticed correlations or connections between studies might be more prosaic than the LBCM structure described above, but it is an LLM use case that has yielded real world results for life sciences players already. The potential benefits of having reasoning machines help in the search for ever more effective treatments cannot be ignored, and we expect adoption of these technologies across the pharmaceutical sector for a wide variety of use cases to be almost explosively rapid.
You can find more views from the DLA Piper team on the topics of AI systems, regulation and the related legal issues on our blog, Technology’s Legal Edge.
If your organisation is deploying AI solutions, you can undertake a free maturity risk assessment using our AI Scorebox tool.
If you’d like to discuss any of the issues discussed in this article, get in touch with Gareth Stokes, Michael Stead or your usual DLA Piper contact.
[1] Not so in this case. The authors can assure you that this entire article has been artisanally hand-crafted by humans. Therefore any errors are our own, and can’t be blamed on machine hallucinations.
[2] There is an excellent examination of this phenomenon, “Emergent Abilities of Large Language Models”, by Wei et al. (https://arxiv.org/abs/2206.07682) which indicates that many properties start to noticeably emerge as the total compute resource required to train the model (a function of architecture, parametric size and training dataset size) is in the 1022 to 1024 floating point operations range.
[3] The word ‘size’ here is doing a lot of heavy lifting, and referring to some combination of parametric size of the model, number of training tokens used to train the model, and thus the computational resources committed to training the model.
[4] A succinct statement of this can be found in this video interview with Sutskever conducted by Dwarkesh Patel: https://youtu.be/Yf1o0TQzry8?t=444 (starting just after 7:25)
[5] For any technically minded readers who might take issue with this framing, the authors accept this is a huge oversimplification of the training by backpropagation, calculation of cost functions and fine tuning by reinforcement learning with human feedback that characterises the training of these models in reality.
[6] By a delightful co-incidence for the subject matter of this section, the ‘longest word’ in English is a whopping 189,819 letters long and is the name of a specific protein. https://en.wikipedia.org/wiki/Longest_word_in_English
[7] For example, https://slator.com/gpt-4-launch-new-use-cases/ and https://seo.ai/blog/how-many-languages-does-chatgpt-support
[8] When accessed directly via API calls, some more recent models will allow between 4,000 and 25,000 tokens of input: https://drewisdope.com/chatgpt-limits-words-characters-tokens/
[9] See https://en.wikipedia.org/wiki/Amino_acid
[10] https://imb.uq.edu.au/article/2017/11/explainer-peptides-vs-proteins-whats-difference#:~:text=Both%20peptides%20and%20proteins%20are,of%20amino%20acids%20than%20proteins
[11] And once again, this monster protein is the same one whose name is so very many letters long in English. https://en.wikipedia.org/wiki/Titin
[12] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5837046/#:~:text=Thus%2C%20if%20a%20single%20representative,%2Dcoding%20genes5%2C6
[13] An explorable map of pathways that makes the London Tube map look sparse can be viewed at https://www.genome.jp/pathway/map01100
[14] The authors readily admit this is a big and entirely untested assumption underpinning this idea.
[15] Much of the detail regarding the biochemical data in this section has been collected by following the source links provided by Kurzgesagt for their recent video ‘The Most Complex Language in the World’ (https://www.youtube.com/watch?v=TYPFenJQciw). Those source links can be found here: https://sites.google.com/view/sources-impossible-machines/ . The video is well worth watching, and indeed some of the thought process for considering the application of LLM structures to biochemistry was inspired when one of the authors was watching this and other educational videos with his children.
[16] See note 3 above.